European foulbrood: Diversity and genotyping written by Victoria Tomkies, manager of the bee health diagnostics laboratory, Fera Science Ltd and Giles Budge, professor at the School of Natural and Environmental Sciences, Newcastle University
European foulbrood control
England and Wales have been deploying a European foulbrood (EFB) control programme over 80 years. The control regime has developed considerably since the early 1940s to incorporate new technologies and improve our understanding of disease spread. Major improvements in control have come with the use of GPS to highlight proximity to previous cases; the use of modelling tools to optimise the rules for risk-based inspections; and the deployment of lateral flow devices to confirm the presence of the disease in the field and enable more rapid treatment. We are all too familiar with using lateral flow devices (LFDs) in recent years and how they can help rapid diagnosis and instant decision making. We are also more familiar with the concept of pathogens, like viruses, having the ability to produce alpha, beta, gamma, omicron variants, and that these variants can sometimes manifest in subtly different disease symptoms. The bacterium that causes EFB, can also have variants, and the geographical distribution of these variants can teach us a great deal about the transmission of this damaging honey bee brood disease.
Measuring the diversity of EFB
The method we use for understanding the diversity of Melissococcus plutonius, the bacterium that causes EFB, is known as Multi Locus Sequence Typing (MLST). This method compares the pattern of genetic code from four genes to allocate each isolate (genetic type) of M. plutonius into a unique sequence type (Haynes et al, 2013). Isolates from all over the world have been tested and allocated a sequence type, allowing us to compare patterns of spread in many different countries. Similar methods are used to help understand the disease transmission for many different human diseases. To date, 39 different sequence types have been identified worldwide, with 28 of these found in the UK. Why is this information important? The first thing a bee inspector might need to do after treating EFB is to consider where the disease may have come from. Imagine how much easier it would be if we could narrow down the potential origin of any new case of EFB? Perhaps this would allow them to focus inspection efforts in a different area. Or maybe provide evidence that we are getting on top of a particular outbreak, or even eradicating it.

Identical sequence types can be used to link outbreaks – or sometimes equally usefully dissimilar sequence types can be used to exclude links between outbreaks. Interestingly, sequence types can be grouped together to form bacterial family groups known as clonal complexes, and these can share biological properties. For example, it has been shown in Japan, that M. plutonius from different clonal complexes can cause more severe disease in laboratory-reared larvae (Nakamura et al, 2016), and that field symptoms of clonal complexes in the UK can cause more severe disease in the field than others (Budge et al, 2014).
Any strain that causes more serious disease will be more easily seen by beekeepers and bee inspectors, and therefore more likely to be reported and treated than those with low virulence, which could lead to more insidious infections. Sequence type information can therefore provide important clues that links outbreaks, and also reveal information about how disease might behave in the field.
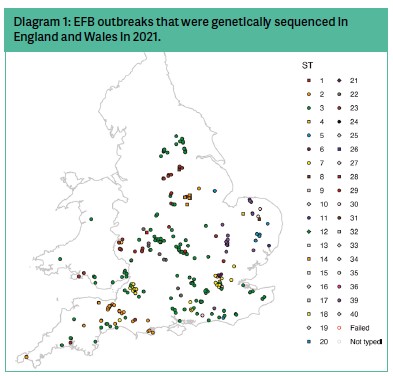
Since 2014, the NBU laboratory at Fera has been taking the majority of positive LFDs from EFB cases across England and Wales and generating sequence-type information. Working with Newcastle University, we have been able to visualise these data through space and time to provide unrivalled resolution about the spread of EFB across England and Wales. Below we share some of the findings from the work and provide specific examples of how understanding diversity can reveal clues about this enigmatic disease. Diagram 1 shows the geographical location of all typed cases from England and Wales in 2021.
Sunrise and sunset genotypes
Ordering our data by time (Diagram 2) allows us to better understand if there are some sequence types of M. plutonius that are becoming more common, perhaps suggesting they are escaping controls. Or perhaps other sequence types that are becoming rarer, or even disappearing altogether suggesting successful control or even complete eradication. These data indicate that sequence types 7 and 39 are becoming more common, and that sequence types 18, 20, 26, 28, 31 and 35 have not been detected in the past two years.
Linking outbreaks
Different sequence types have shown a varying degree of movement across the landscape in recent years. Here we consider two case studies in detail: ST7 and ST39.
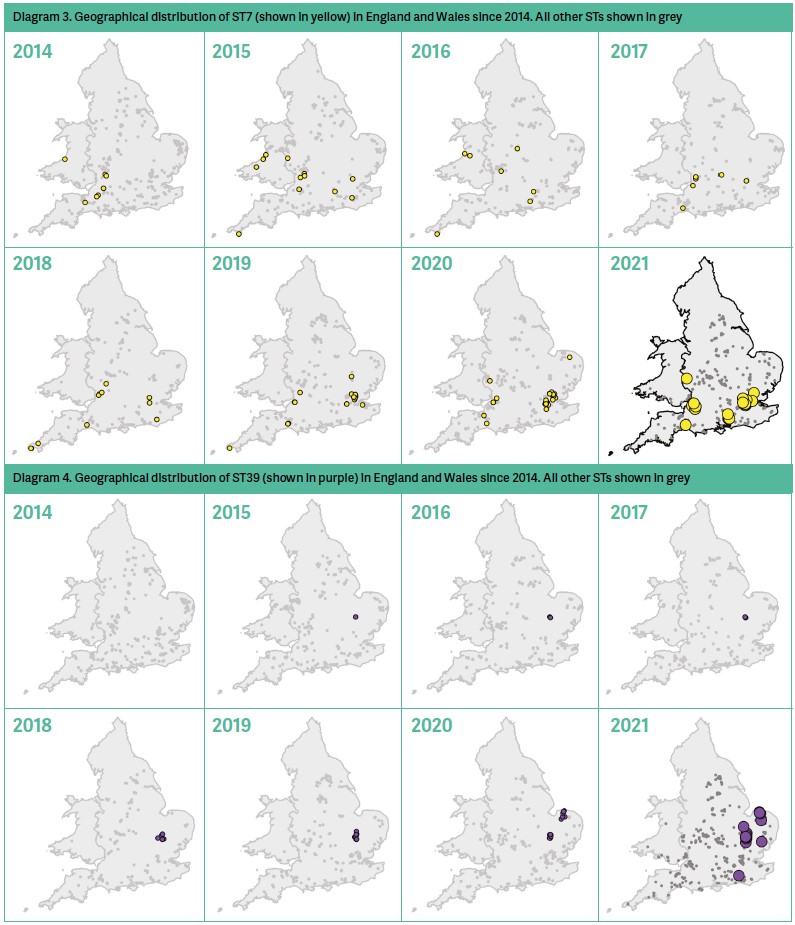
ST7 has been detected in England and Wales since the typing programme started in 2014. Whereas once it was confined to the west (Avon, Somerset and Wales), over time it has been spreading east and is now widespread. It is now the predominant sequence type in Greater London, but remains present in the west, particularly in Avon. Diagram 3 shows the movement of distribution of ST7 from 2014–2021.
ST39 was first identified in Cambridge in 2015. Cases increased each year, but it remained confined to the city and surrounding area for six years. In 2020 the first cases were identified in Norfolk and by 2021 it was also identified in Suffolk and Sussex. Anecdotal evidence from bee inspectors suggests that ST39 presents atypically, with the ‘melted down’ stage rarely seen, and that infection occurs across an apiary by the time the first symptoms show. Interestingly, some sequence types lack the virulence factors that break down the gut (Takamatsu et al, 2016), and it would be interesting to see whether ST39 lacked this virulence factor. Diagram 4 shows the movement of distribution of ST39 from 2014–2021.
Summary
The combination of scientific understanding of the genetic relatedness of M. plutonius adds a string to our bow when trying to improve our understanding of EFB spread. These data are helping us to build a picture of EFB disease at the landscape level and provide information about all the ways through which EFB spreads. We are currently investigating whether some sequence types are better controlled by either shook swarm or colony destruction, and we are developing new methods to provide a more rapid result which we hope will improve disease control. Anything we can do as a community to help prevent the spread of EFB can only be good news for beekeeping across the UK.
REFERENCES
Budge et al (2014). Molecular epidemiology and population structure of the honey bee brood pathogen Melissococcus plutonius. The ISME Journal 8 1588-1597
Haynes E, Helgason T, Young J P W, Thwaites R. and Budge G E (2013). A typing scheme for the honeybee pathogen Melissococcus plutonius allows detection of disease transmission events and a study of the distribution of variants. Environmental Microbiology Reports, 5:525-529
Nakamura et al (2016). Virulence differences among Melissococcus plutonius strains with different genetic backgrounds in Apis mellifera larvae under improved experimental condition Scientific Reports 6:33329 https://doi.org/10.1038/srep33329
Takamatsu D, Morinishi K, Arai R, Sakamoto A, Okura M and Osaki M (2014). Typing of Melissococcus plutonius isolated from European and Japanese honeybees suggests spread of sequence types across borders and between different Apis species. Veterinary Microbiology, 171:221-226
 Victoria Tomkies has worked at Fera Science Ltd (formerly CSL) for more than 20 years. She moved into the National Bee Unit in 2009, originally in research. She has been the diagnostics manager for bee health since 2017.
Victoria Tomkies has worked at Fera Science Ltd (formerly CSL) for more than 20 years. She moved into the National Bee Unit in 2009, originally in research. She has been the diagnostics manager for bee health since 2017.
 Professor Giles Budge is an applied scientist with expertise in pathology, apiculture, molecular biology and crop protection. He has more than 20 years’ experience of applied research in three national organisations and is now at Newcastle University.
Professor Giles Budge is an applied scientist with expertise in pathology, apiculture, molecular biology and crop protection. He has more than 20 years’ experience of applied research in three national organisations and is now at Newcastle University.
Click the logo to download the article.
July 2022





